Derrin Bright
Bachelors Student, Biotechnology
Vellore Institute of Technology, India
Bio: I've always been interested in how biology and technology can work together. My focus lies at that intersection, where I use computational tools and machine learning to explore biological data and understand how diseases function. Using my skills in bioinformatics, I want to help build that bridge between biology and artificial intelligence.
Currently, I am pursuing my undergraduate degree in Biotechnology at Vellore Institute of Technology, Vellore. Recent projects include analyzing transcriptomic data to uncover dysregulated pathways in autoimmune diseases and using molecular docking to visualize how a potential drug compound binds to its target protein, identifying the key interactions that make the binding effective.
I have also worked with computational tools such as AlphaFold for protein structure prediction, GROMACS and AutoDock for molecular simulations, and data analytics platforms like KNIME, alongside machine learning frameworks such as TensorFlow and Keras. Outside of my academic work, I enjoy playing cricket and chess, as well as spending time outdoors.
Core Interests:
- Structural Bioinformatics
- Genomics and Transcriptomics
- Machine Learning in Bioinformatics
Education
Vellore Institute of Technology - Vellore, India
CGPA: 8.69/10

Experience
Boston University - Massachusetts, USA

University of Liverpool - Liverpool, UK

IIT Bombay - Mumbai, India

Ashoka University - Delhi, India

NovaLinks - Vellore, India

Teachnook - Bangalore, India

Bversity - Bangalore, India

Alpha Bio Cell, VIT University - Vellore, India

Skillset















Projects
Identification of Conserved Functional Modules and Unique Hub Genes across Autoimmune Disorders
Tools: GSEA, NetworkAnalyst, R Studio
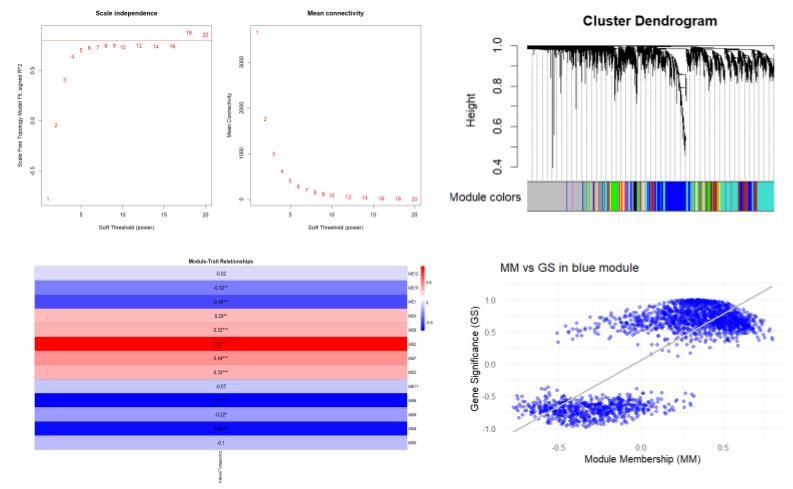
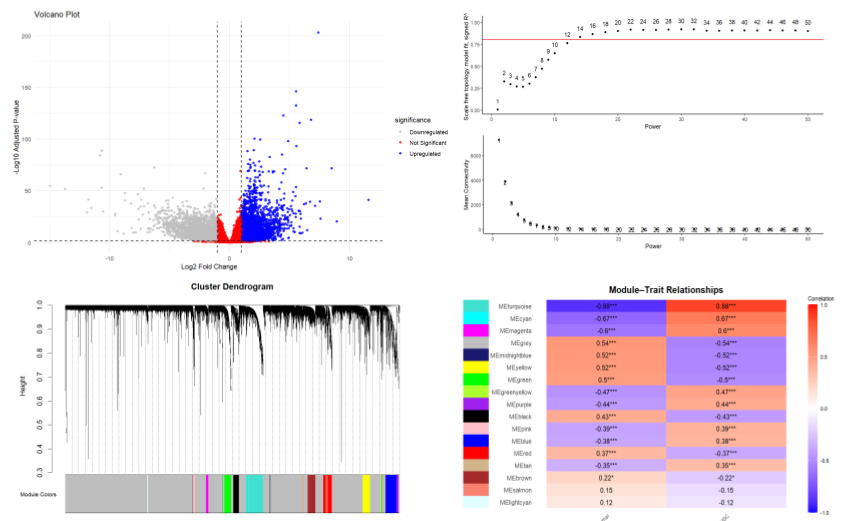
Comparative Transcriptomic Analysis of HPV-Associated Cancers Using Co-Expression Network Modeling
Tools: DESEQ2, WGCNA, Cytoscape, ClusterProfiler
Multi-Modal Genomic Analysis of Beta-Lactam Resistance Mechanisms in Five Clinical E. coli Isolates
Tools: Prokka, Biopython, ResFinder, CARD-RGI, BLAST
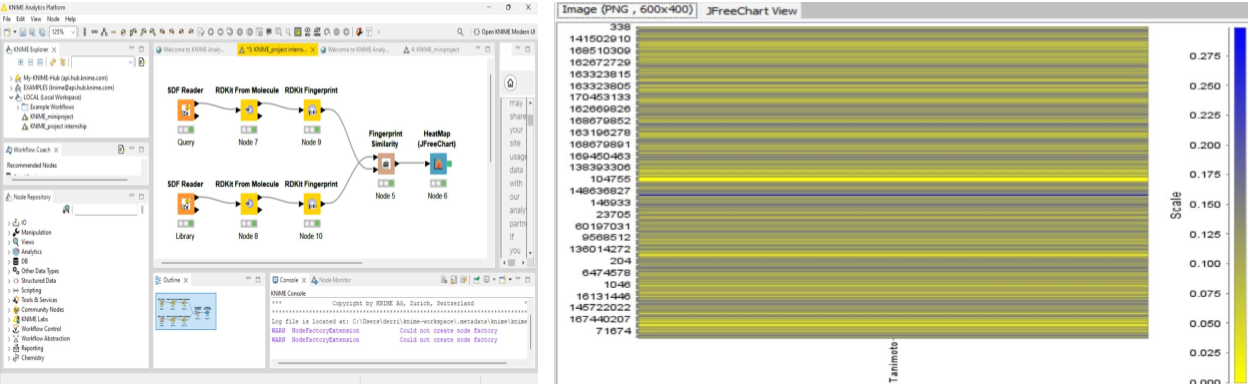
Computational Identification of Antimalarial Leads through Fingerprint Similarity Analysis
Tools: KNIME, RDKit, PubChem, Seaborn
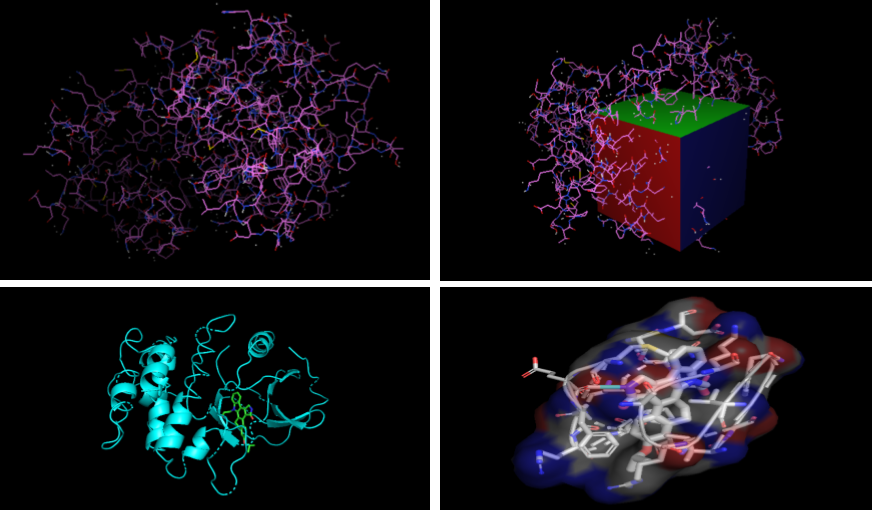
In Silico Analysis of Binding Mode and Affinity for the 3DTC/CEP-6331 Complex using Molecular Docking
Tools: AutoDock Vina, OpenBabel, LigPlot, MGLTools, Avagadro, COACH server
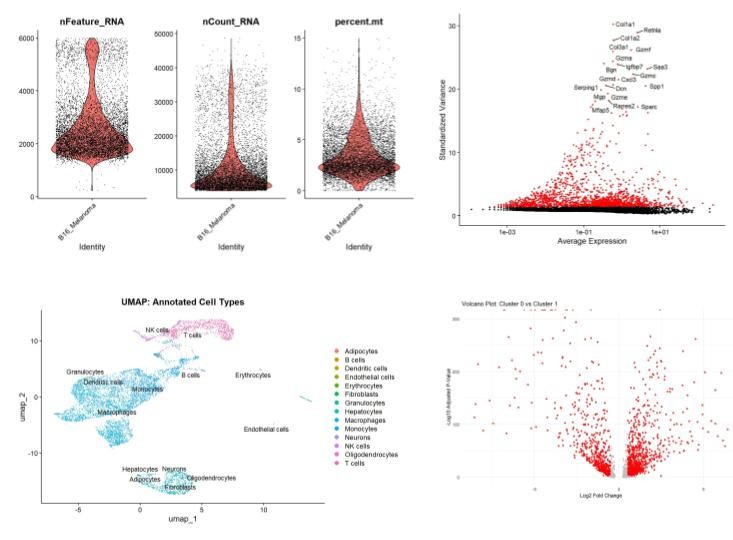
Single-Cell Transcriptomic Profiling of the B16 Melanoma's Microenvironment
Tools: Seurat, SingleR, DESeq2, dplyr
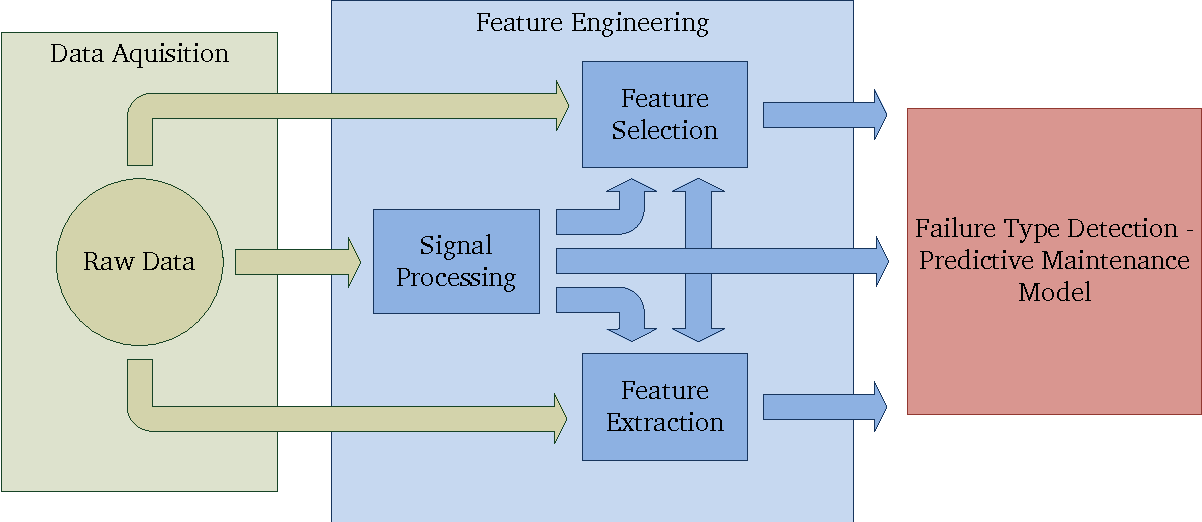
Data-Driven Machine Failure Detection
Tools: Pandas, NumPy, Scikit-learn, Seaborn
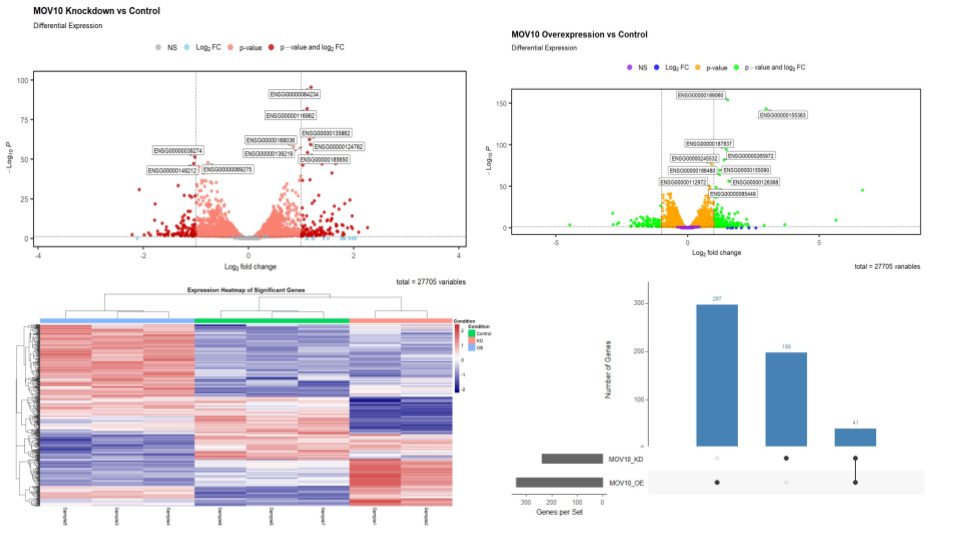
Computational Analysis of Transcriptional Regulation by the MOV10 Protein
Tools: SRA Toolkit, FastQC, Trimmomatic, HISAT2, Subread, DESEQ2, UpSetR, ggVennDiagram, EnhancedVolcano, pheatmap
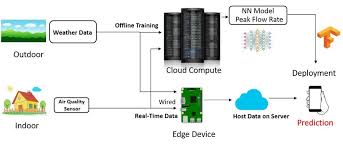
Asthma Risk Prediction
Tools: Pandas, NumPy, Scikit-learn, Seaborn
Notable Achievements
- First runner-up in the Biomimicry Innovation Challenge and Showcase, Vellore Institue of Technology, 2024
- Top 10 Winner of Bio-Inspired Design Fest (BIDFEST) Ideathon, Vellore Institue of Technology, 2024
Certificates
- Machine Learning A-Z, Udemy
- Python for Data Science, AI & Development, Coursera
- Bioinformatic; Bulk RNA-Seq Data Analysis, Udemy
- Advanced Bioinformatics, BioGrademy
- Single-Cell RNA-Seq Data Analysis Using R & Python, Udemy
- Docking and Molecular Dynamics Simulation, Udemy
- Oracle Cloud Infrastructure AI Foundations Associate Certificate, Oracle
- Oracle Cloud Infrastructure Generative AI Professional Certificate, Oracle
Extra-curricular
National Service Scheme, NGO - Vellore, India

Becoming I Foundation, NGO - Vellore, India

Eco Club, Chettinad Vidyashram - Chennai, India

BioSummit, School of Biosciences and Technology - Vellore, India

Contact me
Feel free to reach out via email ; will get back ASAP!